Note: this post has been updated to reflect upgrades to CUDA 12.6, RHEL 9.4, and select packages.

Upgrade RHEL 9 to 9.4
# RHEL 9 to 9.4 # Check the current version of RHEL cat /etc/redhat-release sudo subscription-manager release # List the versions available sudo subscription-manager release --list # Set the release version you want sudo subscription-manager release --set 9.4 # Update sudo dnf update # Confirm the release version cat /etc/redhat-release
EPEL
Extra packages for Enterprise Linux are required for getting Nvidia CUDA and NTFS support to work.
sudo subscription-manager repos --enable codeready-builder-for-rhel-9-$(arch)-rpms sudo dnf install \ https://dl.fedoraproject.org/pub/epel/epel-release-latest-9.noarch.rpm sudo dnf config-manager --enable epel
Docker
# Remove previous installations
sudo yum remove docker \
docker-client \
docker-client-latest \
docker-common \
docker-latest \
docker-latest-logrotate \
docker-logrotate \
docker-engine \
podman \
runc
# Install Docker
sudo yum install -y yum-utils
sudo yum-config-manager --add-repo https://download.docker.com/linux/rhel/docker-ce.repo
sudo yum install docker-ce docker-ce-cli containerd.io docker-buildx-plugin docker-compose-plugin
sudo systemctl start docker
# Add the current user to the docker group
sudo groupadd docker
sudo usermod -aG docker $USERDocumentation for Docker is an excellent place to start. The commands above are there
Nvidia CUDA
While not absolutely required, using a GPU is highly preferred for next-gen -omics projects like scRNA-seq and spatial transcriptomics. GPUs are optimized to run computations in parallel. This is helpful when you are trying to find relationships between tens of thousands of cells, each with tens of thousands of genes.
CUDA, or the Compute Unified Device Architecture, is a technology by Nvidia that allows software to communicate with the GPU through an common API. It is the backbone of what is powering the major advancements in “artificial intelligence” technologies over the last few years.
Getting CUDA to run properly is exactly turnkey “easy to setup and ready to go”. As always, it’s highly recommended to read the official documentation for installing CUDA. Going into the details of the installing CUDA is dependent on a variety of factors, including the Linux distribution, Kernel version, CPU architecture, etc. Below are instructions for my server, and it may likely need to be adjusted for different setups.
Check system requirements
# CUDA-Capable GPU lspci | grep -i nvidia # Supported Version of Linux uname -m && cat /etc/*release # GCC compiler installed gcc --version # Kernal version and CPU architecture uname -srm
Installing CUDA
Once you have confirmed that your system is capable of utilizing CUDA technology, you can proceed to the installation.
# Installing CUDA using the local RPM installer mkdir ~/tmp/cuda && cd ~/tmp/cuda # Download the local installer (4.4 GB) # If your computer does not have internet access, you can download this file on a separate machine and copy it over wget https://developer.download.nvidia.com/compute/cuda/12.6.1/local_installers/cuda-repo-rhel9-12-6-local-12.6.1_560.35.03-1.x86_64.rpm # Kernel headers and development packages sudo dnf install kernel-devel-$(uname -r) kernel-headers-$(uname -r) # Install CUDA repository sudo rpm -i cuda-repo-rhel9-12-6-local-12.6.1_560.35.03-1.x86_64.rpm sudo dnf clean all # Install the cuda-toolkit sudo dnf -y install cuda-toolkit-12-6 # Driver installer sudo dnf -y module install nvidia-driver:open-dkms # GPUDirect Filesystem sudo dnf install nvidia-gds
Note: I am using the newer, open-source driver, which supports “Turning, Ampere, and forward” GPU architectures. If you need legacy support or need to switch between drivers, follow the instructions here.
After the installation is complete, you will be must reboot your computer to complete the Post Install Actions.
# Add CUDA to $PATH in your ~/.bashrc, ~/.bash_profile, ~/.bash_aliases
# For all users, you can create a shell script to load on login: /etc/profile.d/cuda.sh
sudo nano /etc/profile.d/cuda.sh
# Then add this line:
export PATH=/usr/local/cuda-12.6/bin${PATH:+:${PATH}}
# And change permissions
sudo chmod 755 /etc/profile.d/cuda.sh
### If you are using zsh, you can edit:##################
sudo nano /etc/zshenv
# And add the following:
export PATH=/usr/local/cuda-12.6/bin${PATH:+:${PATH}}
#-------------------------------------------------------
# Logout and log back in of the terminal
logout
# Confirm that CUDA has been added to $PATH
echo $PATH | grep cuda
ls -lahg /usr/local
ls -lahg /etc/alternatives | grep cuda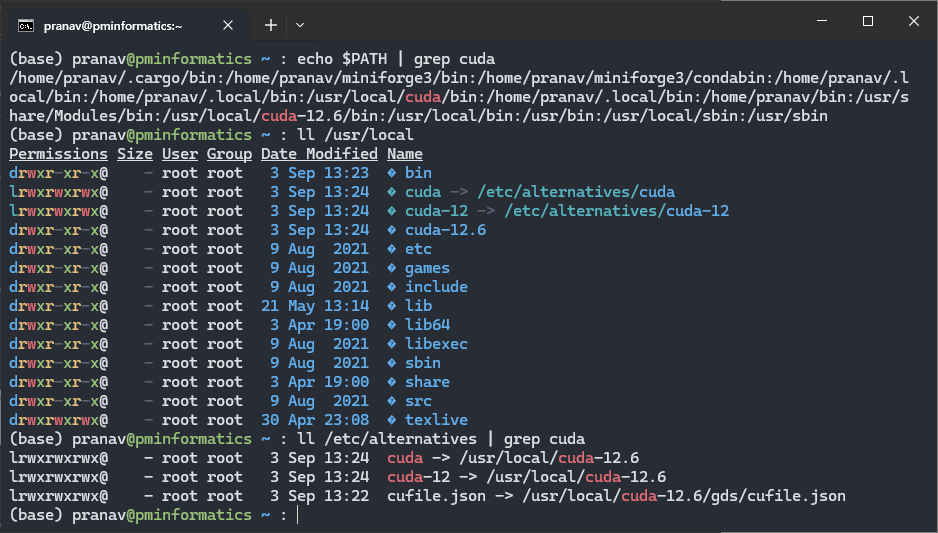
Nvidia uses a chain of symlinks. The TLDR is that you can have multiple versions of CUDA on your computer and switch between them by pointing the symlink to a different version.
# Start the persistance daemon sudo /usr/bin/nvidia-persistenced --verbose
Verify Installation
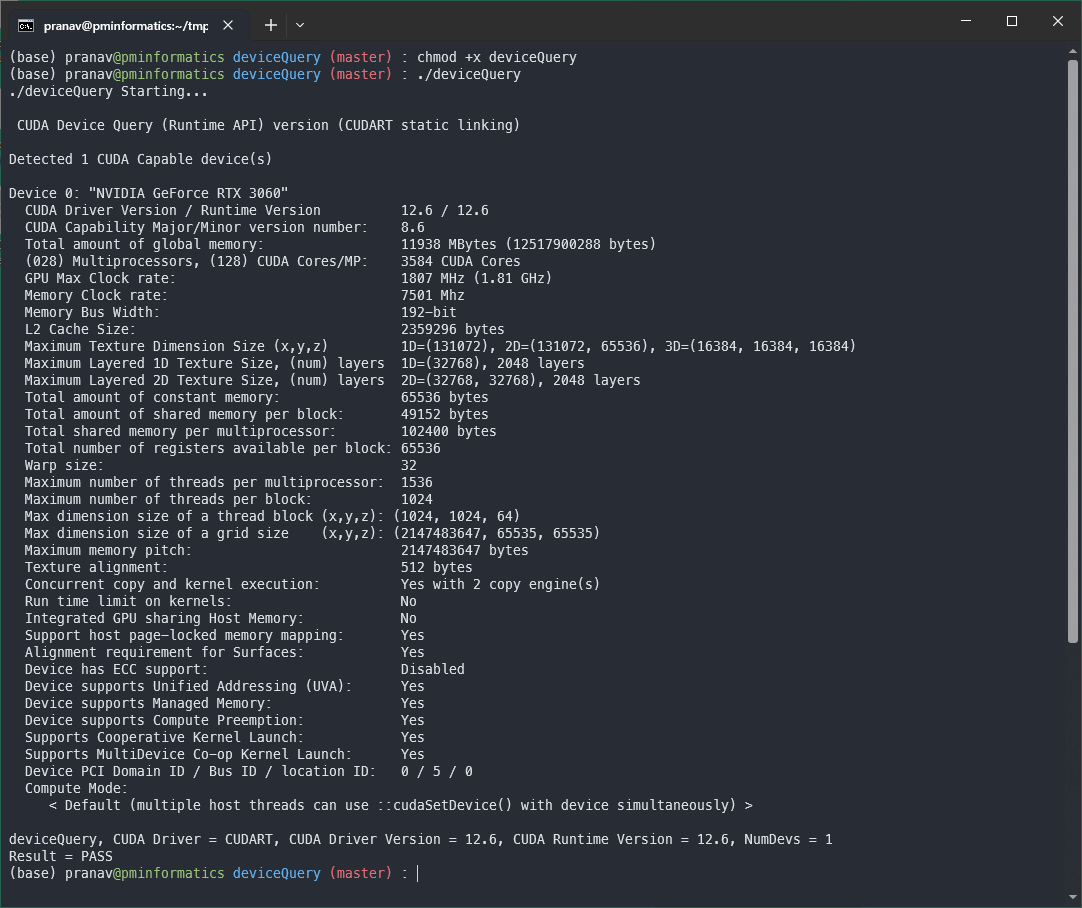
cat /proc/driver/nvidia/version # Device Query using Nvidia CUDA Samples cd ~/tmp/cuda git clone https://github.com/NVIDIA/cuda-samples.git cd ~/tmp/cuda/cuda-samples/Samples/1_Utilities/deviceQuery make chmod +x deviceQuery ./deviceQuery
Nvidia container toolkit
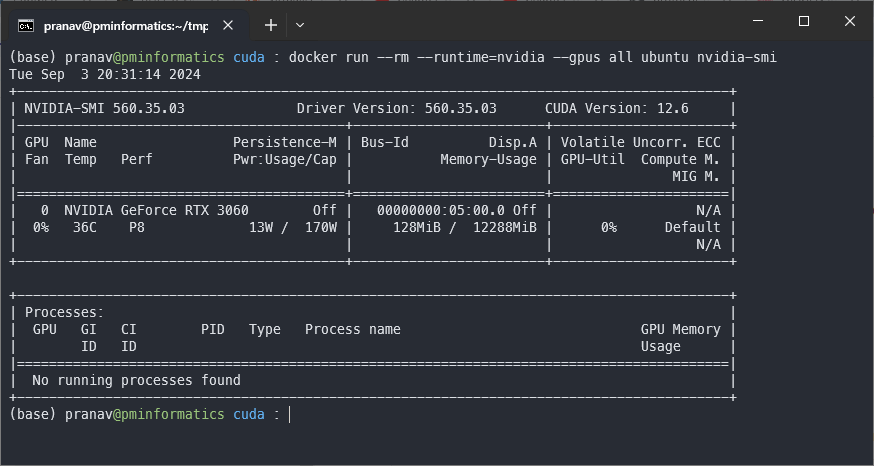
curl -s -L https://nvidia.github.io/libnvidia-container/stable/rpm/nvidia-container-toolkit.repo | \ sudo tee /etc/yum.repos.d/nvidia-container-toolkit.repo sudo yum install -y nvidia-container-toolkit # Tell the container toolkit that we are using docker sudo nvidia-ctk runtime configure --runtime=docker # Restart docker sudo systemctl restart docker # Test your GPU passthrough to a docker container docker run --rm --runtime=nvidia --gpus all ubuntu nvidia-smi
Optional
cd ~/tmp wget --max-redirect=20 -O vscode-linux.rpm "https://code.visualstudio.com/sha/download?build=stable&os=linux-rpm-x64" sudo dnf install ./vscode-linux.rpm wget --max-redirect=20 -O code "https://code.visualstudio.com/sha/download?build=stable&os=cli-alpine-x64" tar -xvf vscode_cli_alpine_x64_cli.tar.gz
Quarto
export QUARTO_VERSION="1.6.32" # replace with the latest version
#Download and install
sudo mkdir -p /opt/quarto/${QUARTO_VERSION}
sudo curl -o quarto.tar.gz -L \
"https://github.com/quarto-dev/quarto-cli/releases/download/v${QUARTO_VERSION}/quarto-${QUARTO_VERSION}-linux-amd64.tar.gz"
sudo tar -zxvf quarto.tar.gz \
-C "/opt/quarto/${QUARTO_VERSION}" \
--strip-components=1
sudo rm quarto.tar.gz
sudo rm /usr/local/bin/quarto
sudo ln -s /opt/quarto/${QUARTO_VERSION}/bin/quarto /usr/local/bin/quarto
# Check installation
quarto checkYadm – Managing Dotfiles
My dot files can be found on my GitHub repository: pranavmishra90/dotfiles.
curl -fLo ~/.local/bin/yadm https://github.com/yadm-dev/yadm/raw/master/yadm && chmod a+x ~/.local/bin/yadm # My dotfiles yadm clone [email protected]:pranavmishra90/dotfiles.git yadm checkout ~
Eza – a modernized replacement for ‘ls’
sudo dnf install eza
Zoxide – an improvement on cd
# Zoxide curl -sSfL https://raw.githubusercontent.com/ajeetdsouza/zoxide/main/install.sh | sh
NTFS Support
If you need access to windows partitions (NTFS) on internal / external hard drives, you need to add the package ntfs-3g
sudo yum install ntfs-3g # Create a mount point mkdir -p /media/pranav/DATA-3 #(my hard drive is called DATA-3) # List hard drives. If you are having difficulty identifying which one, mount the drive using the file browser. You should be able to see something like /dev/sd____ lsblk
Add the following to /etc/fstab, replacing with the correct values from lsblk.
# Internal drives /dev/sda2 /media/pranav/DATA-3 ntfs defaults 0 0
Then, restart the daemon with systemctl daemon-reload
XRDP
# XRDP sudo yum install -y xrdp sudo systemctl enable xrdp sudo systemctl start xrdp echo "gnome-session" > ~/.xsession chmod +x ~/.xsession
Node and NPM
sudo dnf module enable nodejs:20 sudo dnf install nodejs
GitHub Signed Commits
# Download and trust GitHub's GPG key # Trusting at level 4 of 5: "I trust fully" curl -L https://github.com/web-flow.gpg | gpg --import && printf "968479A1AFF927E37D1A566BB5690EEEBB952194:4\n" | gpg --import-ownertrust
Pipx packages
sudo dnf install pipx pipx install pre-commit python-semantic-release
VSCode
cd ~/tmp curl -Lk 'https://code.visualstudio.com/sha/download?build=stable&os=cli-alpine-x64' --output vscode_cli.tar.gz tar -xf vscode_cli.tar.gz # move the binary within your $PATH mv code ~/.local/bin/code # Run as a service code tunnel service install sudo loginctl enable-linger $USER # Start the service code tunnel